Authors: Joshua B. Tenenbaum, Vin de Silva, John C. Langford
The following figures are copied from the original paper.
This work introduces a method called Isomap for dimension reduction specifically on non-linear data. The algorithm composes of three steps:
(1) Find neighbors for each points in input space. To do so, it can be done (a) by retaining K neighbors or (b) by retaining neighbors in a constat radius range for each point. Then, represents neighbors relationship as a weighted graph, in which the weight is the distance between two points.
(2) Compute shortest paths length for all pair points on the neighbors graph. Using the all pair shortest path algorithm like Floyd’s algorithm to detect all pair shortest path lengths.
(3) Perform dimension reduction by applying MDS. Transfer data into low d-dimensions space Y by performing MDS on pair distances calculated in step (2). Like MDS and PCA, the true dimension d in Isomap can be estimated from error loss decreasing in transfered space Y when d increases, and also Isomap can recover true data structure if there are sufficient data.
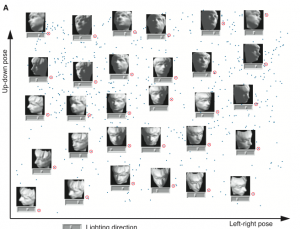
My Opinon: As the experiment result showed above, Isomap finds the structure of data well. However, just authors mentioned, it takes much data to detect the structure of a specific object, and also the time complexity is high due to the step 2 O(n^3) algorithm. I am curious that whether it is useful or not in the real application context.
Go Top